Python is a great programming language, but packaging is one of its weakest points. It is a well-known fact in the community. The process of installing, importing, using, and creating packages has improved a lot over the years, but it's still not on par with newer languages like Go and Rust that learned a lot from the struggles of Python and other mature languages.
In this tutorial, you'll learn everything you need to know about writing, packaging, and distributing your own packages.
How to Write a Python Library
A Python library is a coherent collection of Python modules that is organized as a Python package. In general, that means that all modules live under the same directory and that this directory is on the Python search path.
Let's quickly write a little Python 3 package and illustrate all these concepts.
The Pathology Package
Python 3 has an excellent Path object, which is a huge improvement over Python 2's awkward os.path module. But it's missing one crucial capability—finding the path of the current script. This is very important when you want to locate access files relative to the current script.
In many cases, the script can be installed in any location, so you can't use absolute paths, and the working directory can be set to any value, so you can't use a relative path. If you want to access a file in a sub-directory or parent directory, you must be able to figure out the current script directory.
Here is how you do it in Python:
1 |
import pathlib |
2 |
|
3 |
script_dir = pathlib.Path(__file__).parent.resolve() |
To access a file called 'file.txt' in the 'data' sub-directory of the current script's directory, you can use the following code: print(open(str(script_dir/'data/file.txt').read())
With the pathology package, you have a built-in script_dir method, and you use it like this:
1 |
from pathology.Path import script_dir |
2 |
|
3 |
print(open(str(script_dir()/'data/file.txt').read()) |
4 |
Yep, it's a mouthful. The pathology package is very simple. It derives its own Path class from pathlib's Path and adds a static script_dir() that always returns the path of the calling script.
Here is the implementation:
1 |
import pathlib |
2 |
import inspect |
3 |
|
4 |
class Path(type(pathlib.Path())): |
5 |
@staticmethod |
6 |
def script_dir(): |
7 |
print(inspect.stack()[1].filename) |
8 |
p = pathlib.Path(inspect.stack()[1].filename) |
9 |
return p.parent.resolve() |
Due to the cross-platform implementation of pathlib.Path, you can derive directly from it and must derive from a specific sub-class (PosixPath or WindowsPath). The script_dir resolution uses the inspect module to find the caller and then its filename attribute.
Testing the Pathology Package
Whenever you write something that is more than a throwaway script, you should test it. The pathology module is no exception. Here are the tests using the standard unit test framework:
1 |
import os |
2 |
import shutil |
3 |
from unittest import TestCase |
4 |
from pathology.path import Path |
5 |
|
6 |
|
7 |
class PathTest(TestCase): |
8 |
def test_script_dir(self): |
9 |
expected = os.path.abspath(os.path.dirname(__file__)) |
10 |
actual = str(Path.script_dir()) |
11 |
self.assertEqual(expected, actual) |
12 |
|
13 |
def test_file_access(self): |
14 |
script_dir = os.path.abspath(os.path.dirname(__file__)) |
15 |
subdir = os.path.join(script_dir, 'test_data') |
16 |
if Path(subdir).is_dir(): |
17 |
shutil.rmtree(subdir) |
18 |
os.makedirs(subdir) |
19 |
file_path = str(Path(subdir)/'file.txt') |
20 |
content = '123' |
21 |
open(file_path, 'w').write(content) |
22 |
test_path = Path.script_dir()/subdir/'file.txt' |
23 |
actual = open(str(test_path)).read() |
24 |
|
25 |
self.assertEqual(content, actual) |
The Python Path
Python packages must be installed somewhere on the Python search path to be imported by Python modules. The Python search path is a list of directories and is always available in sys.path. Here is my current sys.path:
1 |
>>> print('\n'.join(sys.path)) |
2 |
|
3 |
/Users/gigi.sayfan/miniconda3/envs/py3/lib/python36.zip |
4 |
/Users/gigi.sayfan/miniconda3/envs/py3/lib/python3.6 |
5 |
/Users/gigi.sayfan/miniconda3/envs/py3/lib/python3.6/lib-dynload |
6 |
/Users/gigi.sayfan/miniconda3/envs/py3/lib/python3.6/site-packages |
7 |
/Users/gigi.sayfan/miniconda3/envs/py3/lib/python3.6/site-packages/setuptools-27.2.0-py3.6.egg |
Note that the first empty line of the output represents the current directory, so you can import modules from the current working directory, whatever it is. You can directly add or remove directories to/from sys.path.
You can also define a PYTHONPATH environment variable, and there are a few other ways to control it. The standard site-packages is included by default, and this is where packages you install using via pip go.
How to Package a Python Library
Now that we have our code and tests, let's package it all into a proper library. Python provides an easy way via the setup module. You create a file called setup.py in your package's root directory.
The setup.py file includes a lot of metadata information such as author, license, maintainers, and other information regarding the package. This is in addition to the packages item, which uses the find_packages() function imported from setuptools to find sub-packages.
Here is the setup.py file of the pathology package:
1 |
from setuptools import setup, find_packages |
2 |
|
3 |
setup(name='pathology', |
4 |
version='0.1', |
5 |
url='https://github.com/the-gigi/pathology', |
6 |
license='MIT', |
7 |
author='Gigi Sayfan', |
8 |
author_email='the.gigi@gmail.com', |
9 |
description='Add static script_dir() method to Path', |
10 |
packages=find_packages(exclude=['tests']), |
11 |
long_description=open('README.md').read(), |
12 |
zip_safe=False) |
Source Distribution Package
A source distribution package refers to an archive file that contains Python packages, modules, and other files that are used for a package release (for example, version 1, 2, and so on). Once the file has been distributed, end users can download and install it on their operating system.
To create a source distribution package (sdist), you run: python setup.py sdist
Let's build a source distribution:
1 |
$ python setup.py sdist |
2 |
running sdist |
3 |
running egg_info |
4 |
creating pathology.egg-info |
5 |
writing pathology.egg-info/PKG-INFO |
6 |
writing dependency_links to pathology.egg-info/dependency_links.txt |
7 |
writing top-level names to pathology.egg-info/top_level.txt |
8 |
writing manifest file 'pathology.egg-info/SOURCES.txt' |
9 |
reading manifest file 'pathology.egg-info/SOURCES.txt' |
10 |
writing manifest file 'pathology.egg-info/SOURCES.txt' |
11 |
warning: sdist: standard file not found: should have one of README, README.rst, README.txt |
12 |
|
13 |
running check |
14 |
creating pathology-0.1 |
15 |
creating pathology-0.1/pathology |
16 |
creating pathology-0.1/pathology.egg-info |
17 |
copying files to pathology-0.1... |
18 |
copying setup.py -> pathology-0.1 |
19 |
copying pathology/__init__.py -> pathology-0.1/pathology |
20 |
copying pathology/path.py -> pathology-0.1/pathology |
21 |
copying pathology.egg-info/PKG-INFO -> pathology-0.1/pathology.egg-info |
22 |
copying pathology.egg-info/SOURCES.txt -> pathology-0.1/pathology.egg-info |
23 |
copying pathology.egg-info/dependency_links.txt -> pathology-0.1/pathology.egg-info |
24 |
copying pathology.egg-info/not-zip-safe -> pathology-0.1/pathology.egg-info |
25 |
copying pathology.egg-info/top_level.txt -> pathology-0.1/pathology.egg-info |
26 |
Writing pathology-0.1/setup.cfg |
27 |
creating dist |
28 |
Creating tar archive |
29 |
removing 'pathology-0.1' (and everything under it) |
The warning is because I used a non-standard README.md file. It's safe to ignore. The command above will create an archive file with the default format for the current operating system. For Unix systems, a gzipped tar file will be generated, under the dist directory:
1 |
$ ls -la dist |
2 |
total 8 |
3 |
drwxr-xr-x 3 gigi.sayfan gigi.sayfan 102 Apr 18 21:20 .
|
4 |
drwxr-xr-x 12 gigi.sayfan gigi.sayfan 408 Apr 18 21:20 .. |
5 |
-rw-r--r-- 1 gigi.sayfan gigi.sayfan 1223 Apr 18 21:20 pathology-0.1.tar.gz
|
If you are using Windows, a zip file is generated.
You can also specify other additional file formats using the format option as follows.
1 |
python setup.py sdist --formats=gztar,zip |
For example, the above command will generate a gzipped tarball and a zip file.
The different formats available are:
-
zip: .zip -
gztar: .tar.gz -
bztar: .tar.bz2 -
xztar: .tar.xz -
ztar: .tar.Z -
tar: .tar
Binary Distribution
To create a binary distribution, called a wheel, you run: python setup.py bdist_wheel
And here is a binary distribution:
1 |
$ python setup.py bdist_wheel |
2 |
running bdist_wheel |
3 |
running build |
4 |
running build_py |
5 |
creating build |
6 |
creating build/lib |
7 |
creating build/lib/pathology |
8 |
copying pathology/__init__.py -> build/lib/pathology |
9 |
copying pathology/path.py -> build/lib/pathology |
10 |
installing to build/bdist.macosx-10.7-x86_64/wheel |
11 |
running install |
12 |
running install_lib |
13 |
creating build/bdist.macosx-10.7-x86_64 |
14 |
creating build/bdist.macosx-10.7-x86_64/wheel |
15 |
creating build/bdist.macosx-10.7-x86_64/wheel/pathology |
16 |
copying build/lib/pathology/__init__.py -> build/bdist.macosx-10.7-x86_64/wheel/pathology |
17 |
copying build/lib/pathology/path.py -> build/bdist.macosx-10.7-x86_64/wheel/pathology |
18 |
running install_egg_info |
19 |
running egg_info |
20 |
writing pathology.egg-info/PKG-INFO |
21 |
writing dependency_links to pathology.egg-info/dependency_links.txt |
22 |
writing top-level names to pathology.egg-info/top_level.txt |
23 |
reading manifest file 'pathology.egg-info/SOURCES.txt' |
24 |
writing manifest file 'pathology.egg-info/SOURCES.txt' |
25 |
Copying pathology.egg-info to build/bdist.macosx-10.7-x86_64/wheel/pathology-0.1-py3.6.egg-info |
26 |
running install_scripts |
27 |
creating build/bdist.macosx-10.7-x86_64/wheel/pathology-0.1.dist-info/WHEEL |
The pathology package contains only pure Python modules, so a universal package can be built. If your package includes C extensions, you'll have to build a separate wheel for each platform:
1 |
$ ls -la dist |
2 |
total 16 |
3 |
drwxr-xr-x 4 gigi.sayfan gigi.sayfan 136 Apr 18 21:24 .
|
4 |
drwxr-xr-x 13 gigi.sayfan gigi.sayfan 442 Apr 18 21:24 .. |
5 |
-rw-r--r-- 1 gigi.sayfan gigi.sayfan 2695 Apr 18 21:24 pathology-0.1-py3-none-any.whl
|
6 |
-rw-r--r-- 1 gigi.sayfan gigi.sayfan 1223 Apr 18 21:20 pathology-0.1.tar.gz
|
For a deeper dive into the topic of packaging Python libraries, check out How to Write Your Own Python Packages.
How to Distribute a Python Package
Python has a central package repository called PyPI (Python Packages Index). PyPI makes it easy to manage different versions of packages. For example, if a user needs to install a specific package version, pip knows where to look for it.
When you install a Python package using pip, it will download the package from PyPI (unless you specify a different repository). To distribute our pathology package, we need to upload it to PyPI and provide some extra metadata PyPI requires. The steps are:
- Upgrade your pip version.
- Create an account on PyPI (just once).
- Register your package.
- Upload your package.
Upgrade Your pip Version
Ensure you have the latest version of pip installed in your operating system. To upgrade pip, issue the following command
1 |
python3 -m pip install --upgrade pip |
Create an Account
You can create an account on the PyPI website. Then create a .pypirc file in your home directory:
1 |
[distutils] |
2 |
index-servers=pypi |
3 |
|
4 |
[pypi] |
5 |
repository = https://pypi.python.org/pypi |
6 |
username = the_gigi |
For testing purposes, you can add a pypitest index server to your .pypirc file:
1 |
[distutils] |
2 |
index-servers= |
3 |
pypi
|
4 |
pypitest
|
5 |
|
6 |
[pypitest] |
7 |
repository = https://testpypi.python.org/pypi |
8 |
username = the_gigi |
9 |
|
10 |
[pypi] |
11 |
repository = https://pypi.python.org/pypi |
12 |
username = the_gigi |
Register Your Package
If this is the first release of your package, you need to register it with PyPI. Use the register command of setup.py. It will ask you for your password. Note that I point it to the test repository here:
1 |
$ python setup.py register -r pypitest |
2 |
running register |
3 |
running egg_info |
4 |
writing pathology.egg-info/PKG-INFO |
5 |
writing dependency_links to pathology.egg-info/dependency_links.txt |
6 |
writing top-level names to pathology.egg-info/top_level.txt |
7 |
reading manifest file 'pathology.egg-info/SOURCES.txt'
|
8 |
writing manifest file 'pathology.egg-info/SOURCES.txt'
|
9 |
running check |
10 |
Password: |
11 |
Registering pathology to https://testpypi.python.org/pypi |
12 |
Server response (200): OK |
Upload Your Package
Now that the package is registered, we can upload it. I recommend using twine, which is more secure. Install it as usual using pip install twine. Then upload your package using twine and provide your password (redacted below):
1 |
$ twine upload -r pypitest -p <redacted> dist/* |
2 |
Uploading distributions to https://testpypi.python.org/pypi |
3 |
Uploading pathology-0.1-py3-none-any.whl |
4 |
[================================] 5679/5679 - 00:00:02 |
5 |
Uploading pathology-0.1.tar.gz |
6 |
[================================] 4185/4185 - 00:00:01 |
The package is now available on the PyPI official site, as shown below.
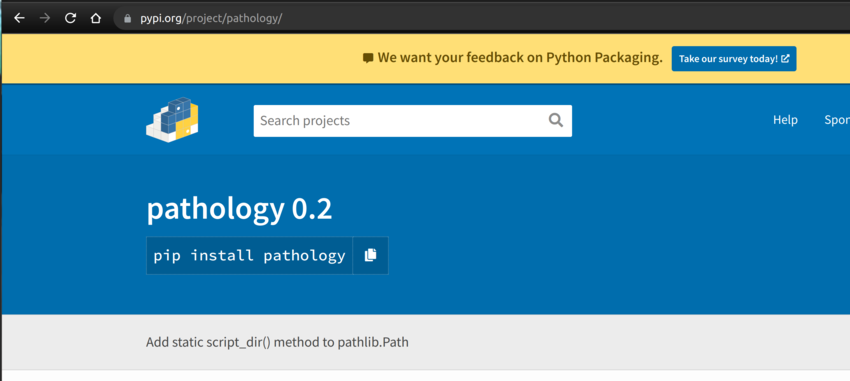


To install it with pip, simply issue the following command:
1 |
pip install pathology
|
For a deeper dive into the topic of distributing your packages, check out How to Share Your Python Packages.
Conclusion
In this tutorial, we went through the fully fledged process of writing a Python library, packaging it, and distributing it through PyPI. At this point, you should have all the tools to write and share your libraries with the world.
This post has been updated with contributions from Esther Vaati. Esther is a software developer and writer for Envato Tuts+.










