When talking about neural networks and deep learning, we usually think about feed-forward neural networks and supervised learning. To be more precise, we are focused on neural networks that have input and output data available to them during the learning process. Based on this information, this kind of neural networks change their weights and are able to learn how to solve a certain problem. However, there are other types of learning and we will explore neural networks that are using these other approaches.
Namely, we are going to get familiar with unsupervised learning. This type of learning gained popularity in the past couple of years. Neural networks that use this type of learning get only input data and based on that they generate some form of output. The correct answers are not known during the learning process and neural networks try to figure out patterns in the data on their own. A result of this approach is that we usually have some kind of clustering or classification of data. Self Organizing Maps, or SOMs for short, are using this approach.
This bundle of e-books is specially crafted for beginners.
Everything from Python basics to the deployment of Machine Learning algorithms to production in one place.
Become a Machine Learning Superhero TODAY!
In the previous article, we got familiar with the main concepts of Self-Organizing Maps and in this one, we will dive even deeper. We explored how they utilize a different type of learning than we got a chance to see during our trip through the world of artificial neural networks – unsupervised learning. This is the type of learning in which the network doesn’t get the expected result for a certain input, but it got to figure out inner data relationships on its own. Self-Organizing Maps (SOM) use this approach for clustering and classification purposes and they are quite good at it. In this article, we cover:
1. Self-Organizing Maps (SOM) Architecture
Even though the early concepts for this type of network can be traced back to 1981, they were developed and formalized in 1992 by Teuvo Kohonen, a professor of the Academy of Finland. In an essence, they are using vector quantization to detect patterns in multidimensional data and represent them in many lower-dimensional spaces – usually one or two dimensions, but we will get into more details later.
However, it is important to take a fresh perspective on these networks and forget standard neuron/connection and weights concepts. These networks are using the same terms but they have a different meaning in their world. Let’s take a look.
There is a reason why these networks are called maps. They are sheet-like neural networks, whose neurons are activated by various patterns or classes of patterns in input signals. Take a look at the picture below:

Here we can see a simple self-organizing map structure. We are having two input neurons, which essentially present features in our dataset. This also means that our input data can be represented by three-dimensional vectors. Above them, we can see so-called map neurons. The goal of these neurons is to present data received on input neurons as two-dimensional data. Meaning, that in this example self-organizing map uses unsupervised learning to cluster that three-dimensional data into a two-dimensional representation.
Of course, we can have any number of dimensions in our input data and any number of dimensions for our output (mapping) data. It is important to notice that each map neuron is connected to each input neuron and that map neurons are not connected to each other. This makes every map neuron oblivious to what values their neighbors have.

Each connection still has a weight attached to it, but they are not used in the same way as in feedforward networks. Basically, you can see that weights are now representing the relationship of the mapping neuron with the input vector. Every map neuron can be identified by unique i, j coordinate and weights on their connections are updated based on the values on the input data, but more on that later.
Now you can see why it is important to take a fresh perspective on this type of network. Even though they are using the same terms, like neurons, connections and weights their meaning is completely different. Apart from that, you can see that their structure is much simpler than the structure of the other feed-forward neural networks. This is causing the learning process to be different as well. So, let’s see how these networks learn.
2. Learning Process of Self-Organizing Maps (SOM)
As we mentioned previously, self-organizing maps use unsupervised learning. This type of learning is also called competitive learning, and we will see in a second why. The first step in the learning process of self-organizing maps is the initialization of all weights on connections.
After that, a random sample from the dataset is used as an input to the network. The network then calculates weights of which neurons are most like the input data (input vector). For this purpose, this formula is used:
where n is the number of connections (weights). The map neuron with the best result is called Best Matching Unit or BMU. In an essence, this means that the input vector can be represented with this mapping neuron. Now, the self-organizing maps are not just calculating this point during the learning process, but they also try to make it “closer” to the received input data.
This means that weights on this connection are updated in a manner that the calculated distance is even smaller. Still, that is not the only thing that is done. The weights of neighbors of BMU are also modified so they are closer to this input vector too. This is how the whole map is ‘pulled’ toward this point. For this purpose, we have to know the radius of the neighbors that will be updated. This radius is initially large, but it is reduced in every iteration (epoch). So, the next step in training self-organizing maps is actually calculating mentioned radius value. The following formula is applied:
where t is the current iteration, σo is the radius of the map. The λ in the formula is defined like this:
where k is the number of iterations. This formula utilizes exponential decay, making the radius smaller as the training goes on, which was the initial goal. In a nutshell, this means that every iteration through the data will bring relevant points closer to the input data. The self-organizing map is fine-tuned in this way.
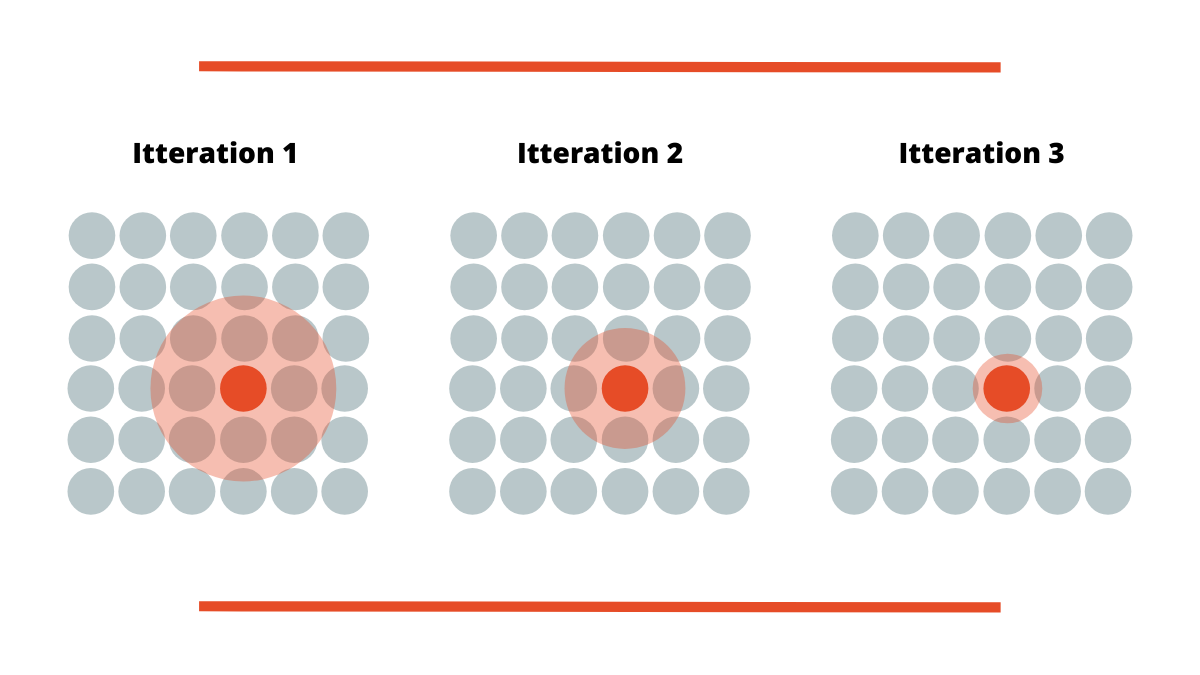
When the radius of the current iteration is calculated weights of all neurons within the radius are updated. The closer the neuron is to the BMU the more its weights are changed. This is achieved by using this formula:
This is the main learning formula, and it has a few important points that should be discussed. The first one is L(t) which represents the learning rate. Similarly to the radius formula, it is utilizing exponential decay and it is getting smaller in every iteration:
Apart from that, we mentioned that the weight of the neuron will be more modified if that neuron is closer to the BMU. In the formula, that is handled with the Θ(t). This value is calculated like this:
Apparently, if the neuron is closer to the BMU, distBMU is smaller, and with that Θ(t) value is closer to 1. This means that the value of the weight of such neuron will be more changed. This whole procedure is repeated several times.

To sum it up, these are the most important steps in the self-organizing map learning process:
- Weight initialization
- The input vector is selected from the dataset and used as an input for the network
- BMU is calculated
- The radius of neighbors that will be updated is calculated
- Each weight of the neurons within the radius are adjusted to make them more like the input vector
- Steps from 2 to 5 are repeated for each input vector of the dataset
Of course, there are a lot of variations of the equations presented used in the learning process of self-organizing maps. In fact, a lot of research has been done trying to get to the optimal values for the number of iterations, the learning rate, and the neighborhood radius. The inventor, Teuvo Kohonen, suggested that this learning process should be split into two phases. During the first phase, the learning rate would be reduced from 0.9 to 0.1 and the neighborhood radius from half the diameter of the lattice to the immediately surrounding nodes.
In the second phase, the learning rate would be further reduced from 0.1 to 0.0. However, there would be double or more iterations in the second phase and the neighborhood radius value should remain fixed at 1, meaning the BMU only. This means that the first phase would be used for learning and the second phase would be used for fine-tuning.
3. Existing Implementations of Self-Organizing Maps (SOM)
The last implementation in the list – MiniSOM is one of the most popular ones. It is a minimalistic, Numpy based implementation of the Self-Organizing Maps and it is very user-friendly. It can be installed using pip:
pip install minisomor using the downloaded setup:
python setup.py installAs mentioned, the usage of this library is quite easy and straightforward. In general, all you have to do is create an object of SOM class, and define its size, size of the input, learning rate, and radius (sigma). After that, you can use one of the two options for training that this implementation provides – train_batch or train_random. The first one uses samples in order in which is recorded in the data set, while the second one shuffles through the samples. Here is an example:
from minisom import MiniSom
som = MiniSom(6, 6, 4, sigma=0.5, learning_rate=0.5)
som.train_random(data, 100)In this example, 6×6 Self-Organizing Map is created, with the 4 input nodes (because data set in this example is having 4 features). Learning rate and radius (sigma) are both initialized to 0.5. Then Self-Organizing Map is trained with input data for 100 iterations using train_random.
4. Implementation with Python and Tensorflow
For this implementation, a low-level API of TensorFlow is used. Here you can find a quick guide on how to quickly install it and how to start working with it. In general, the low-level API of this library is used for the implementation. So, let’s check out the code:
import tensorflow as tf
import numpy as np
class SOM(object):
def __init__(self, x, y, input_dim, learning_rate, radius, num_iter=111):
#Initialize properties
self._x = x
self._y = y
self._learning_rate = float(learning_rate)
self._radius = float(radius)
self._num_iter = num_iter
self._graph = tf.Graph()
#Initialize graph
with self._graph.as_default():
#Initializing variables and placeholders
self._weights = tf.Variable(tf.random_normal([x*y, input_dim]))
self._locations = self._generate_index_matrix(x, y)
self._input = tf.placeholder("float", [input_dim])
self._iter_input = tf.placeholder("float")
#Calculating BMU
input_matix = tf.stack([self._input for i in range(x*y)])
distances = tf.sqrt(tf.reduce_sum(tf.pow(tf.subtract(self._weights, input_matix), 2), 1))
bmu = tf.argmin(distances, 0)
#Get BMU location
mask = tf.pad(tf.reshape(bmu, [1]), np.array([[0, 1]]))
size = tf.cast(tf.constant(np.array([1, 2])), dtype=tf.int64)
bmu_location = tf.reshape(tf.slice(self._locations, mask, size), [2])
#Calculate learning rate and radius
decay_function = tf.subtract(1.0, tf.div(self._iter_input, self._num_iter))
_current_learning_rate = tf.multiply(self._learning_rate, decay_function)
_current_radius = tf.multiply(self._radius, decay_function)
#Adapt learning rate to each neuron based on position
bmu_matrix = tf.stack([bmu_location for i in range(x*y)])
bmu_distance = tf.reduce_sum(tf.pow(tf.subtract(self._locations, bmu_matrix), 2), 1)
neighbourhood_func = tf.exp(tf.negative(tf.div(tf.cast(bmu_distance, "float32"), tf.pow(_current_radius, 2))))
learning_rate_matrix = tf.multiply(_current_learning_rate, neighbourhood_func)
#Update all the weights
multiplytiplier = tf.stack([tf.tile(tf.slice(
learning_rate_matrix, np.array([i]), np.array([1])), [input_dim])
for i in range(x*y)])
delta = tf.multiply(
multiplytiplier,
tf.subtract(tf.stack([self._input for i in range(x*y)]), self._weights))
new_weights = tf.add(self._weights, delta)
self._training = tf.assign(self._weights, new_weights)
#Initilize session and run it
self._sess = tf.Session()
initialization = tf.global_variables_initializer()
self._sess.run(initialization)
def train(self, input_vects):
for iter_no in range(self._num_iter):
for input_vect in input_vects:
self._sess.run(self._training,
feed_dict={self._input: input_vect,
self._iter_input: iter_no})
self._centroid_matrix = [[] for i in range(self._x)]
self._weights_list = list(self._sess.run(self._weights))
self._locations = list(self._sess.run(self._locations))
for i, loc in enumerate(self._locations):
self._centroid_matrix[loc[0]].append(self._weights_list[i])
def map_input(self, input_vectors):
return_value = []
for vect in input_vectors:
min_index = min([i for i in range(len(self._weights_list))],
key=lambda x: np.linalg.norm(vect – self._weights_list[x]))
return_value.append(self._locations[min_index])
return return_value
def _generate_index_matrix(self, x,y):
return tf.constant(np.array(list(self._iterator(x, y))))
def _iterator(self, x, y):
for i in range(x):
for j in range(y):
yield np.array([i, j])That is quite a lot of code, so let’s dissect it into smaller chunks and explain what each piece means. The majority of the code is in the constructor of class which, similar to the MiniSOM implementation, takes dimensions of the Self-Organizing Map, input dimensions, radius and learning rate as input parameters.
4.1 Initialization
The first thing that is done is the initialization of all the fields with the values that are passed into the class constructor:
##Initialize properties
self._x = x
self._y = y
self._learning_rate = float(learning_rate)
self._radius = float(radius)
self._num_iter = num_iter
self._graph = tf.Graph()
Note that we created the TensorFlow graph as a _graph field. In the next part of the code, we essentially add operations to this graph and initialize our Self-Organizing Map. If you need more information on how TensorFlows graphs and sessions work, you can find it here. Anyway, the first step that needs to be done is to initialize variables and placeholders:
#Initializing variables and placeholders
self._weights = tf.Variable(tf.random_normal([x*y, input_dim]))
self._locations = self._generate_index_matrix(x, y)
self._input = tf.placeholder("float", [input_dim])
self._iter_input = tf.placeholder("float")Basically, we created _weights as a randomly initialized tensor. In order to easily manipulate the neurons matrix of indexes is created – _locations. They are generated by using _generate_index_matrix, which looks like this:
def _generate_index_matrix(self, x,y):
return tf.constant(np.array(list(self._iterator(x, y))))
def _iterator(self, x, y):
for i in range(x):
for j in range(y):
yield np.array([i, j])Also, notice that _input (input vector) and _iter_input (iteration number, which is used for radius calculations) are defined as placeholders. This is due to the fact that this information is filled during the training phase, not the construction phase. Once all variables and placeholders are initialized, we can start with the Self-Organizing Map learning process algorithm.
4.2 BMU Calculations
Firstly, BMU is calculated and it’s location is determined:
#Calculating BMU
input_matix = tf.stack([self._input for i in range(x*y)])
distances = tf.sqrt(tf.reduce_sum(tf.pow(tf.subtract(self._weights, input_matix), 2), 1))
bmu = tf.argmin(distances, 0)
#Get BMU location
mask = tf.pad(tf.reshape(bmu, [1]), np.array([[0, 1]]))
size = tf.cast(tf.constant(np.array([1, 2])), dtype=tf.int64)
bmu_location = tf.reshape(tf.slice(self._locations, mask, size), [2])
The first part basically calculates the Euclidean distances between all neurons and the input vector. Don’t get confused by the first line of this code. In essence, this input sample vector is repeated and matrix is created to be used for calculations with weights tensor. Once distances are calculated, the index of the BMU is returned. This index is used, in the second part of the gist, to get the BMU location. We relied on the slice function for this. Once that is done, we need to calculate values for learning rate and radius for the current iteration. That is done like this:
#Adapt learning rate to each neuron based on position
bmu_matrix = tf.stack([bmu_location for i in range(x*y)])
bmu_distance = tf.reduce_sum(tf.pow(tf.subtract(self._locations, bmu_matrix), 2), 1)
neighbourhood_func = tf.exp(tf.negative(tf.div(tf.cast(bmu_distance, "float32"), tf.pow(_current_radius, 2))))
learning_rate_matrix = tf.multiply(_current_learning_rate, neighbourhood_func)The first matrix of BMU location value is created. Then of the neuron to the BMU is calculated. After that, the so-called neighbourhood_func is created. This function is basically defining how the weight of concrete neurons will be changed.
4.3 Update Weights
Finally, the weights are updated accordingly and the TensorFlow session is initialized and run:
#Update all the weights
multiplytiplier = tf.stack([tf.tile(tf.slice(
learning_rate_matrix, np.array([i]), np.array([1])), [input_dim])
for i in range(x*y)])
delta = tf.multiply(
multiplytiplier,
tf.subtract(tf.stack([self._input for i in range(x*y)]), self._weights))
new_weightages = tf.add(self._weights, delta)
self._training = tf.assign(self._weights, new_weightages)
#Initilize session and run it
self._sess = tf.Session()
initialization = tf.global_variables_initializer()
self._sess.run(initialization)
Apart from the _generate_index_matrix function that you saw previously, this class has also two important functions – train and map_input. The first one, as its name suggests, is used to train the Self-Organizing Map with proper input. Here is how that function looks like:
def train(self, input_vects):
for iter_no in range(self._num_iter):
for input_vect in input_vects:
self._sess.run(self._training,
feed_dict={self._input: input_vect,
self._iter_input: iter_no})
self._centroid_matrix = [[] for i in range(self._x)]
self._weights_list = list(self._sess.run(self._weights))
self._locations = list(self._sess.run(self._locations))
for i, loc in enumerate(self._locations):
self._centroid_matrix[loc[0]].append(self._weights_list[i])Essentially, we have just run a defined number of iterations on passed input data. For that, we used _training operation that we created during class construction. Notice that here placeholders for iteration number and input sample are filled. That is how we run created sessions with correct data.
The second function that this class has is map_input. This function is mapping defined input samples to the correct output. Here is how it looks like:
def map_input(self, input_vectors):
return_value = []
for vect in input_vectors:
min_index = min([i for i in range(len(self._weights_list))],
key=lambda x: np.linalg.norm(vect – self._weights_list[x]))
return_value.append(self._locations[min_index])
return return_value4.4 Usage
In the end, we got a Self-Organizing Map with a pretty straightforward API that can be easily used. In the next article, we will use this class to solve one real-world problem. To sum it up, it can be used something like this:
from somtf import SOM
som = SOM(6, 6, 4, 0.5, 0.5, 100)
som.train(data)Conclusion
In this article, we learned how to implement a Self-Organizing map algorithm using TensorFlow. We used flexibility of the lower level API so to get in even more details of their learning process and got comfortable with it. To sum it up, we applied all theoretical knowledge that we learned in the previous article. Apart from that, we saw how we can use already available Self-Organizing implementations, namely MiniSOM. Next step would be using this implementation to solve some real-world problems, which we will do in the future.
Thank you for reading!

Nikola M. Zivkovic
Nikola M. Zivkovic is the author of books: Ultimate Guide to Machine Learning and Deep Learning for Programmers. He loves knowledge sharing, and he is an experienced speaker. You can find him speaking at meetups, conferences, and as a guest lecturer at the University of Novi Sad.





Trackbacks/Pingbacks