Interactive C++ for Data Science
Interactive C++ for Data Science
In our previous blog post “Interactive C++ with Cling” we mentioned that exploratory programming is an effective way to reduce the complexity of the problem. This post will discuss some applications of Cling developed to support data science researchers. In particular, interactively probing data and interfaces makes complex libraries and complex data more accessible to users. We aim to demonstrate some of Cling’s features at scale; Cling’s eval-style programming support; projects related to Cling; and show interactive C++/CUDA.
Eval-style programming
A Cling instance can access itself through its runtime. The example creates a
cling::Value to store the execution result of the incremented variable i.
That mechanism can be used further to support dynamic scopes extending the name
lookup at runtime.
[cling]$ #include <cling/Interpreter/Value.h>
[cling]$ #include <cling/Interpreter/Interpreter.h>
[cling]$ int i = 1;
[cling]$ cling::Value V;
[cling]$ gCling->evaluate("++i", V);
[cling]$ i
(int) 2
[cling]$ V
(cling::Value &) boxes [(int) 2]
V “boxes” the expression result providing extended lifetime if necessary.
The cling::Value can be used to communicate expression values from the
interpreter to compiled code.
[cling]$ ++i
(int) 3
[cling]$ V
(cling::Value &) boxes [(int) 2]
This mechanism introduces a delayed until runtime evaluation which enables some features increasing the dynamic look and feel of the C++ language.
The ROOT data analysis package
The main tool for storage, research and visualization of scientific data in the field of high energy physics (HEP) is the specialized software package ROOT. ROOT is a set of interconnected components that assist scientists from data storage and research to their visualization when published in a scientific paper. ROOT has played a significant role in scientific discoveries such as gravitational waves, the great cavity in the Pyramid of Cheops, the discovery of the Higgs boson by the Large Hadron Collider. For the last 5 years, Cling has helped to analyze 1 EB physical data, serving as a basis for over 1000 scientific publications, and supports software run across a distributed million CPU core computing facility.
ROOT uses Cling as a reflection information service for data serialization. The C++ objects are stored in a binary format, vertically. The content of a loaded data file is made available to the users and C++ objects become a first class citizen.
A central component of ROOT enabled by Cling is eval-style programming. We use this in HEP to make it easy to inspect and use C++ objects stored by ROOT. Cling enables ROOT to inject available object names into the name lookup when a file is opened:
[root] ntuple->GetTitle()
error: use of undeclared identifier 'ntuple'
[root] TFile::Open("tutorials/hsimple.root"); ntuple->GetTitle() // #1
(const char *) "Demo ntuple"
[root] gFile->ls();
TFile** tutorials/hsimple.root Demo ROOT file with histograms
TFile* tutorials/hsimple.root Demo ROOT file with histograms
OBJ: TH1F hpx This is the px distribution : 0 at: 0x7fadbb84e390
OBJ: TNtuple ntuple Demo ntuple : 0 at: 0x7fadbb93a890
KEY: TH1F hpx;1 This is the px distribution
[...]
KEY: TNtuple ntuple;1 Demo ntuple
[root] hpx->Draw()
The ROOT framework injects additional names to the name lookup on two stages.
First, it builds an invalid AST by marking the occurrence of ntuple (#1), then
it is transformed into
gCling->EvaluateT</*return type*/void>("ntuple->GetTitle()", /*context*/);
On the next stage, at runtime, ROOT opens the file, reads its preambule and
injects the names via the external name lookup facility in clang. The
transformation becomes more complex if ntuple->GetTitle() takes arguments.
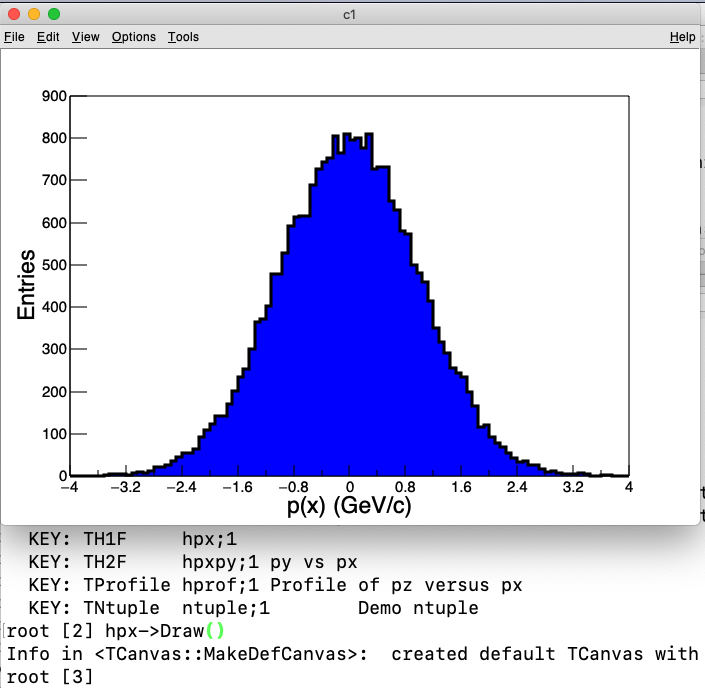
Figure 1. Interactive plot of the px distribution read from a root file.
C++ in Notebooks
Section Author: Sylvain Corlay, QuantStack
The Jupyter Notebook technology allows users to create and share documents that contain live code, equations, visualizations and narrative text. It enables data scientists to easily exchange ideas or collaborate by sharing their analyses in a straight-forward and reproducible way. Language agnosticism is a key design principle for the Jupyter project, and the Jupyter frontend communicates with the kernel (the part of the infrastructure that runs the code) through a well-specified protocol. Kernels have been developed for dozens of programming languages, such as R, Julia, Python, Fortran (through the LLVM-based LFortran project).
Jupyter’s official C++ kernel relies on Xeus, a C++ implementation of the kernel protocol, and Cling. An advantage of using a reference implementation for the kernel protocol is that a lot of features come for free, such as rich mime type display, interactive widgets, auto-complete, and much more.
Rich mime-type rendering for user-defined types can be specified by providing
an overload of mime_bundle_repr for the said type, which is picked up by
argument dependent lookup.
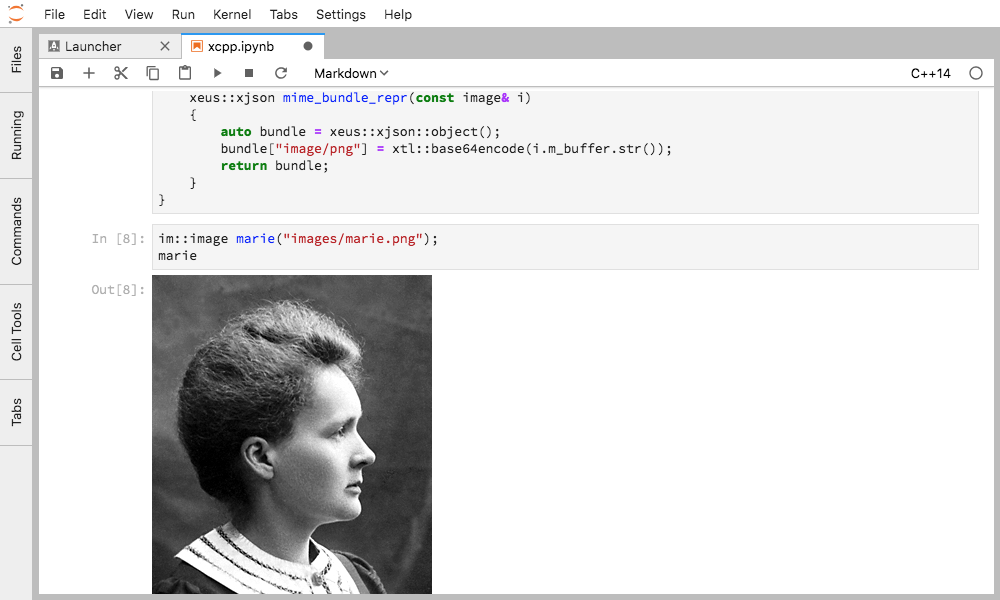
Figure 2. Inline rendering of images in JupyterLab for a user-defined image type.
Possibilities with rich mime type rendering are endless, such as rich display of dataframes with HTML tables, or even mime types that are rendered in the front-end with JavaScript extensions.
An advanced example making use of rich rendering with Mathjax is the SymEngine symbolic computing library.
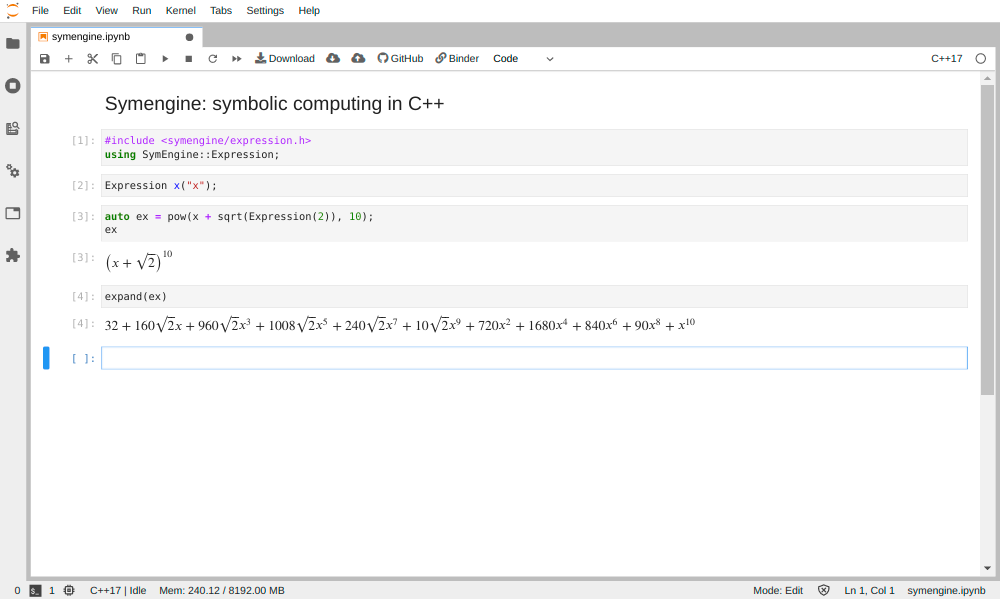
Figure 3. Using rich mime type rendering in Jupyter with the Symengine package.
Xeus-cling comes along with an implementation of the Jupyter widgets protocol which enables bidirectional communication with the backend.
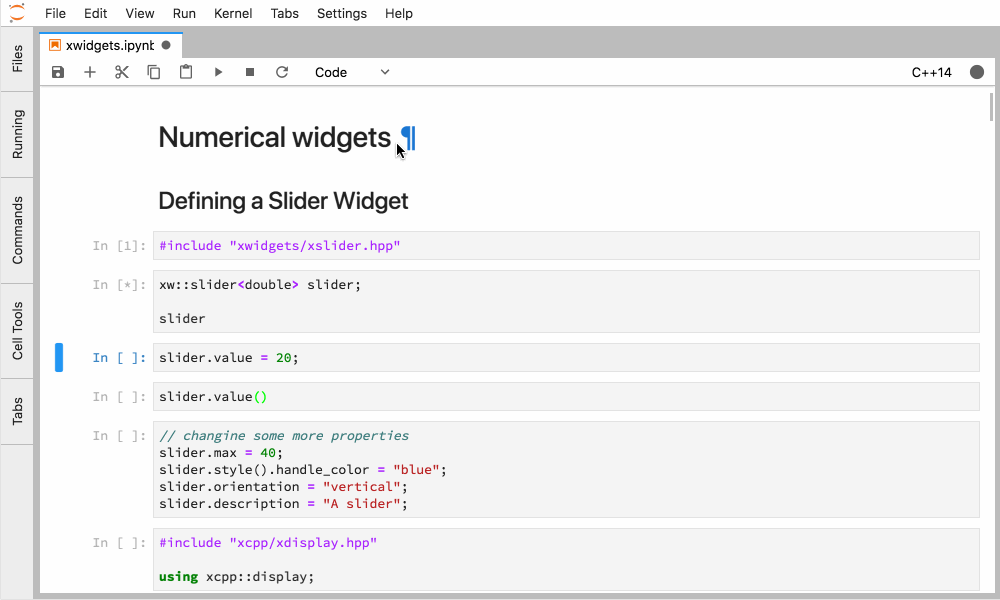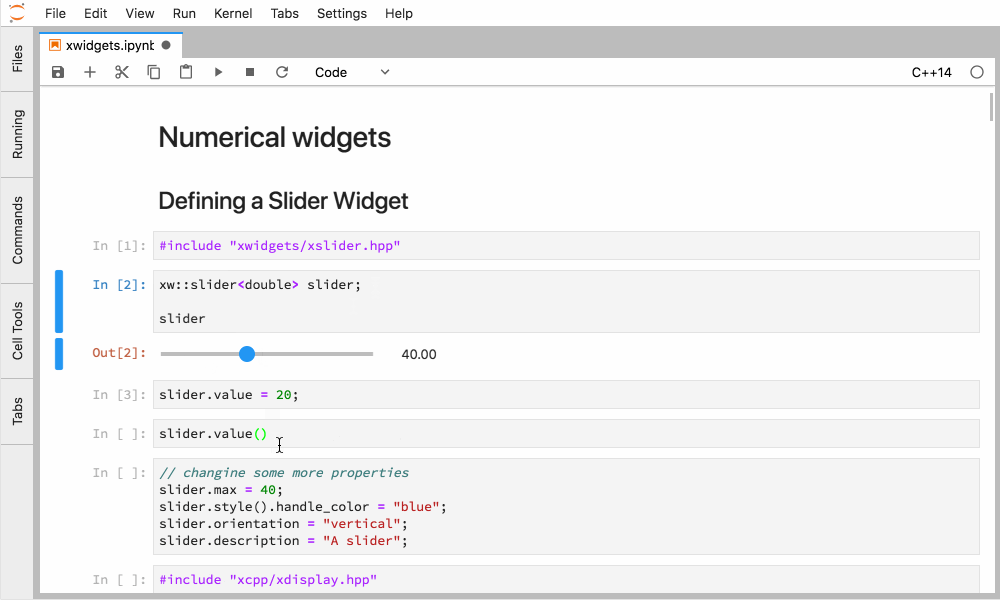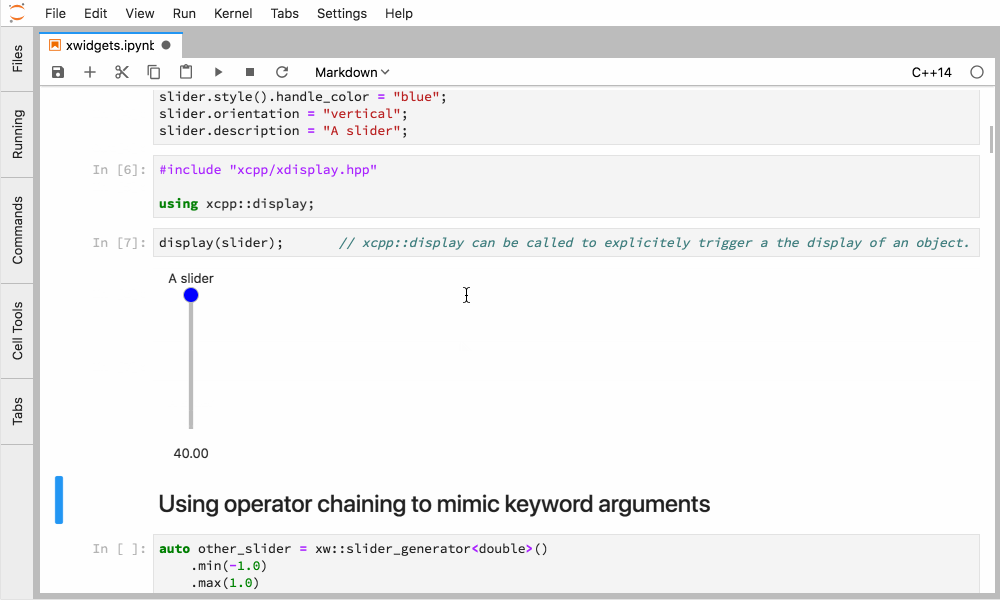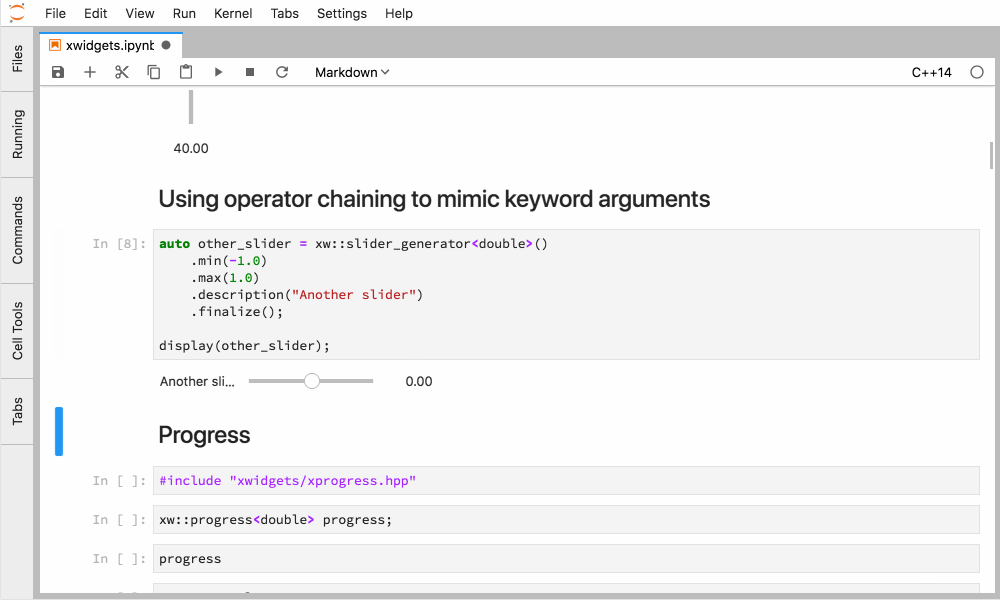
Figure 4. Interactive widgets in the JupyterLab with the C++ kernel.
More complex widget libraries have been enabled through this framework like xleaflet.
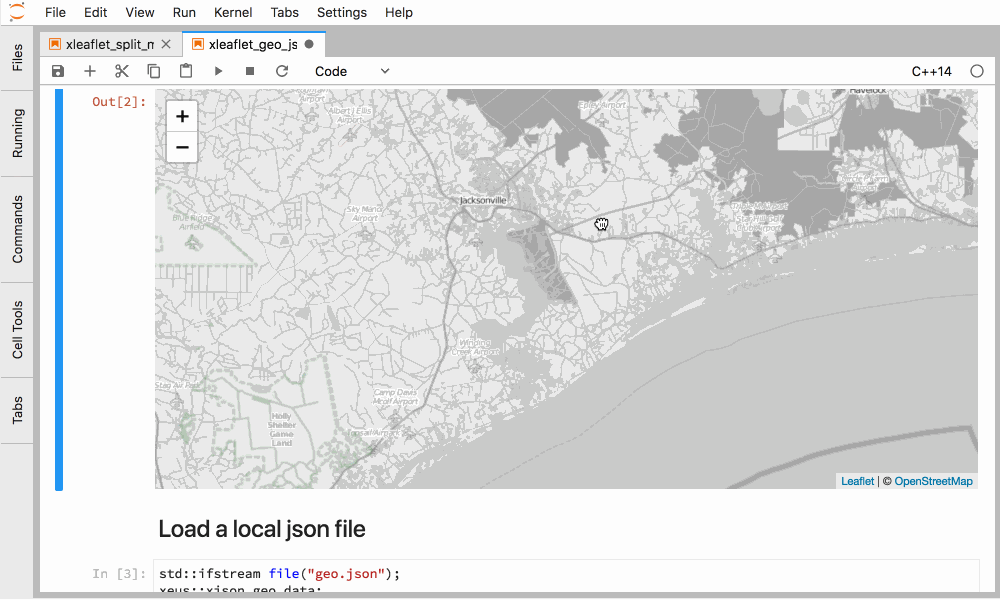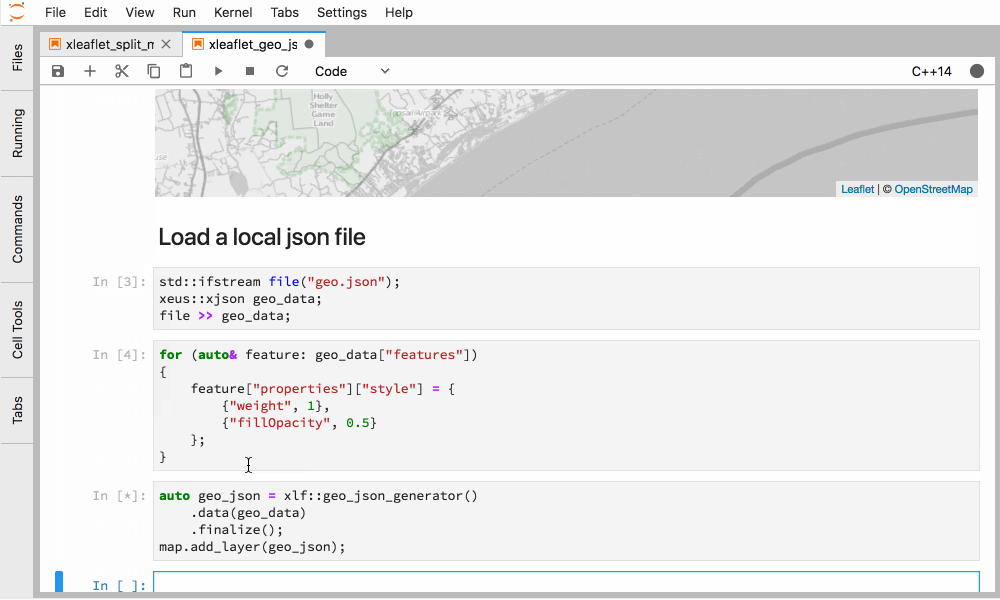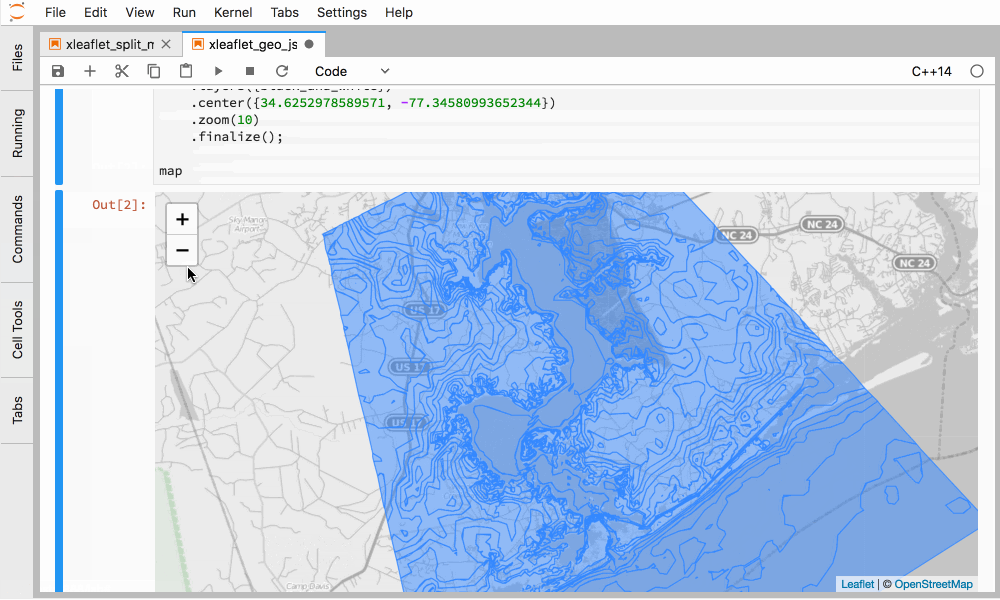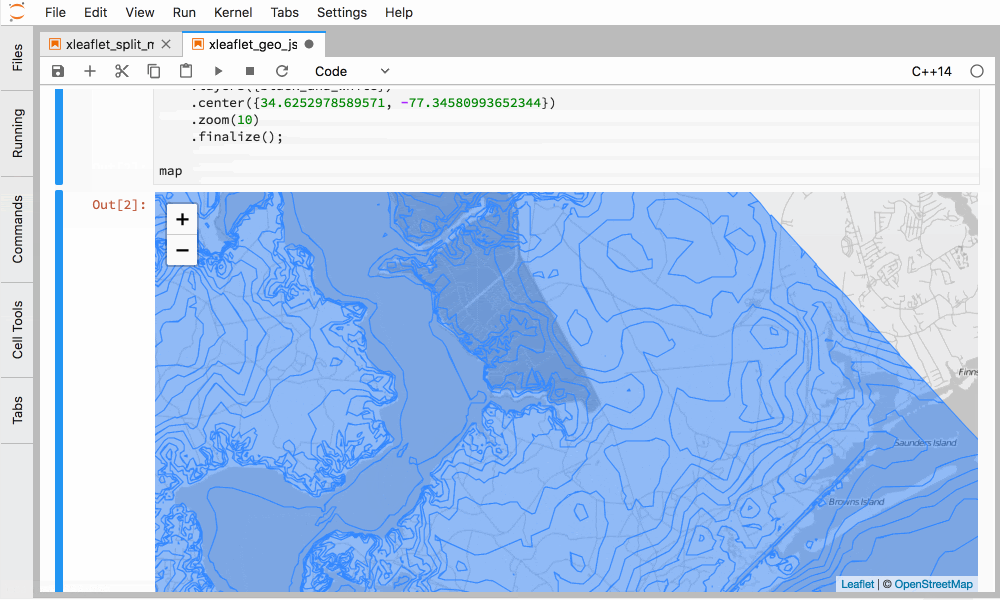
Figure 5. Interactive GIS in C++ in JupyterLab with xleaflet.
Other features include rich HTML help for the standard library and third-party packages:
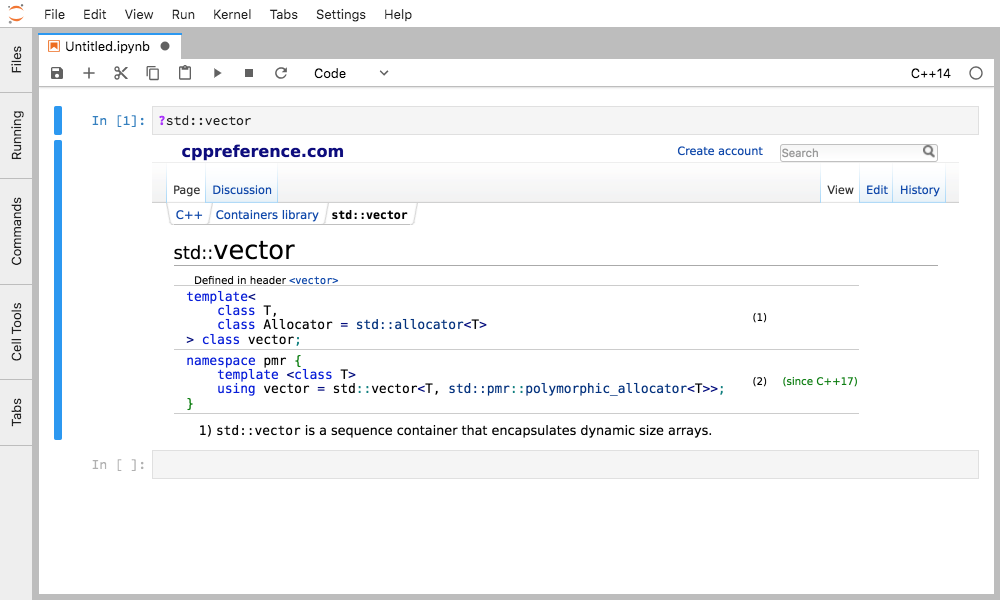
Figure 6. Accessing cppreference for std::vector from JupyterLab by typing `?std::vector`.
The Xeus and Xeus-cling kernels were recently incorporated as subprojects to Jupyter, and are governed by its code of conduct and general governance.
Planned future developments for the xeus-cling kernel include: adding support
for the Jupyter console interface, through an implementation of the Jupyter
is_complete message, currently lacking; adding support for cling
“dot commands” as Jupyter magics; and supporting the new debugger protocol that
was recently added to the Jupyter kernel protocol, which will enable the use of
the JupyterLab visual debugger with the C++ kernel.
Another tool that brings interactive plotting features to xeus-cling is xvega, which is at an early stage of development, produces vega charts that can be displayed in the notebook.
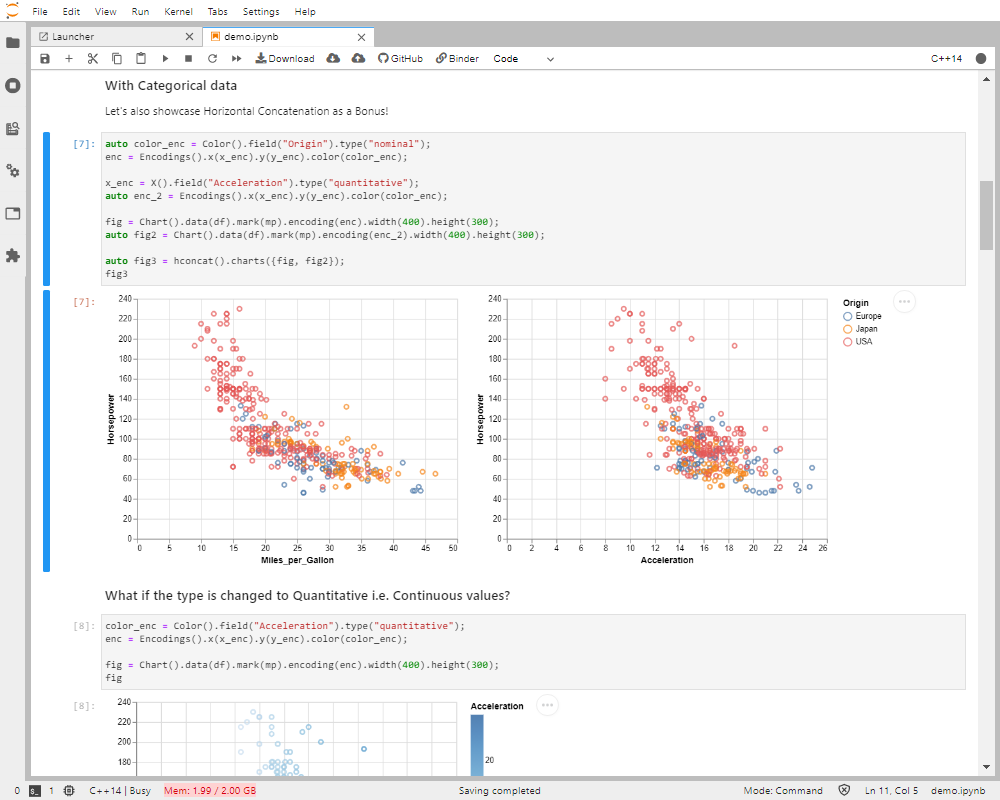
Figure 7. The xvega plotting library in the xeus-cling kernel.
CUDA C++
Section Author: Simeon Ehrig, HZDR
The Cling CUDA extension brings the workflows of interactive C++ to GPUs without losing performance and compatibility to existing software. To execute CUDA C++ Code, Cling activates an extension in the compiler frontend to understand the CUDA C++ dialect and creates a second compiler instance that compiles the code for the GPU.
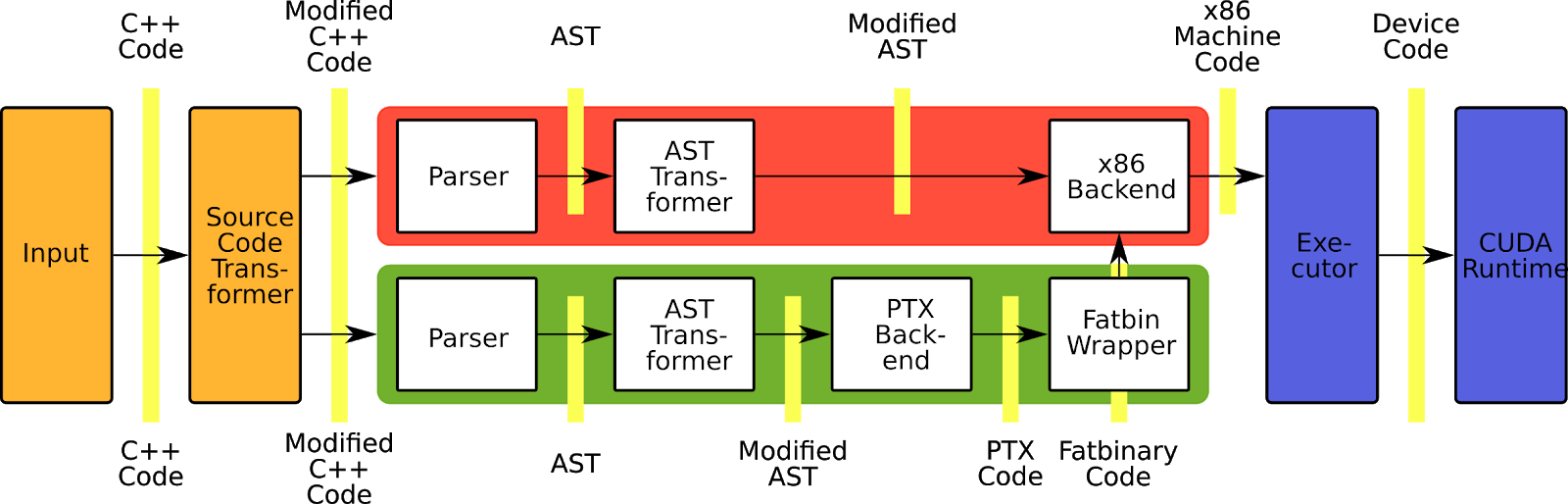
Figure 8. CUDA/C++ information flow in Cling.
Like the normal C++ mode, the CUDA C++ mode uses AST transformation to enable interactive CUDA C++ or special features as the Cling print system. In contrast to the normal Cling compiler pipeline used for the host code, the device compiler pipeline does not use all the transformations of the host pipeline. Therefore, the device pipeline has some special transformation.
[cling] #include <iostream>
[cling] #include <cublas_v2.h>
[cling] #pragma cling(load "libcublas.so") // link a shared library
// set parameters
// allocate memory
// ...
[cling] __global__ void init(float *matrix, int size){
[cling] ? int x = blockIdx.x * blockDim.x + threadIdx.x;
[cling] ? if (x < size)
[cling] ? matrix[x] = x;
[cling] ? }
[cling]
[cling] // launching a function direct in the global space
[cling] init<<<blocks, threads>>>(d_A, dim*dim);
[cling] init<<<blocks, threads>>>(d_B, dim*dim);
[cling]
[cling] cublasSgemm(handle, CUBLAS_OP_N, CUBLAS_OP_N, dim, dim, dim, &alpha, d_A, dim, d_B, dim, &beta, d_C, dim);
[cling] cublasGetVector(dim*dim, sizeof(h_C[0]), d_C, 1, h_C, 1);
[cling] cudaGetLastError()
(cudaError_t) (cudaError::cudaSuccess) : (unsigned int) 0
Like the normal C++ mode, the CUDA mode can be used in a Jupyter Notebook.
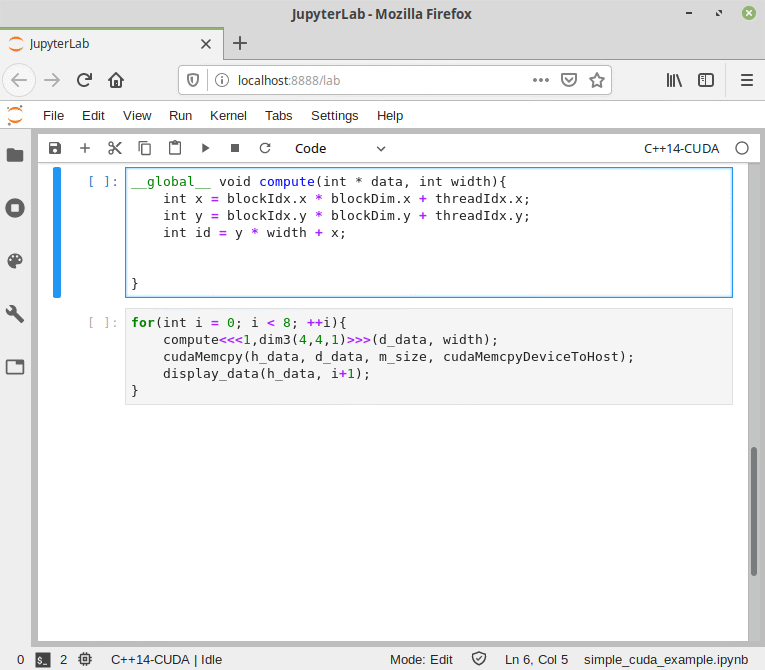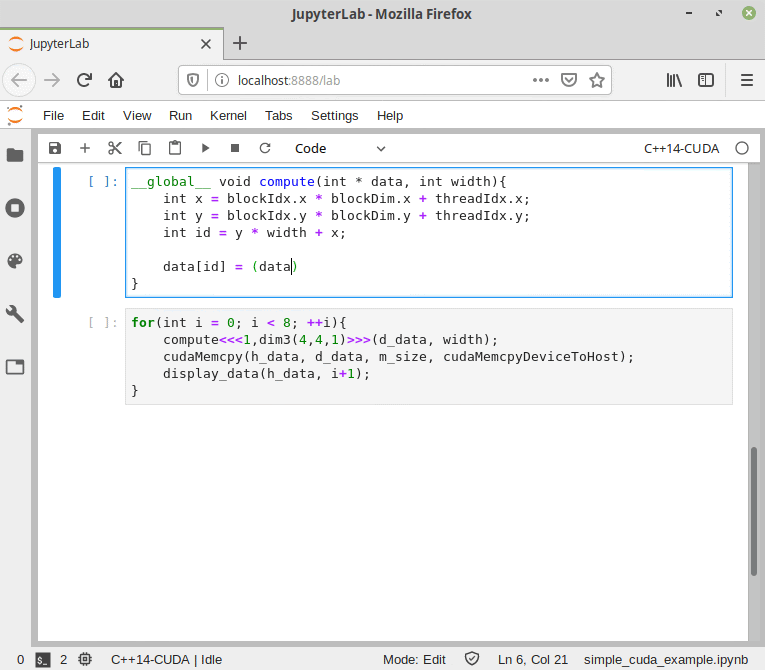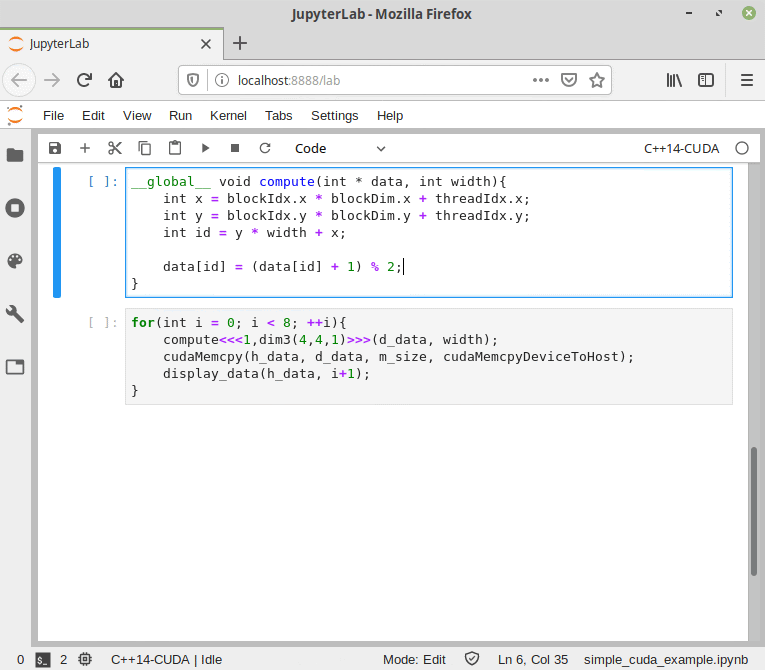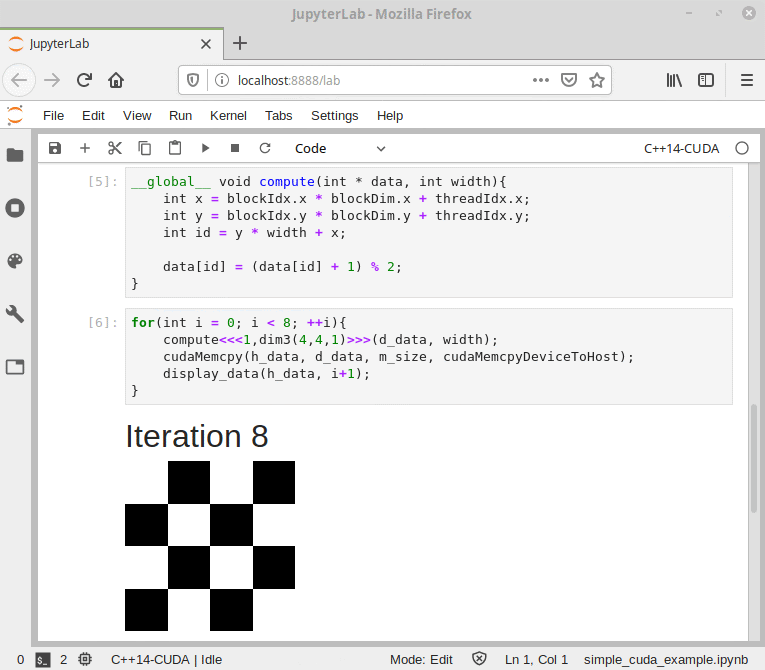
Figure 9. CUDA/C++ information flow in Cling.
A special property of Cling in CUDA mode is that the Cling application becomes a
normal CUDA application at the time of the first CUDA API call. This enables the
CUDA SDK with Cling. For example, you can use the CUDA profiler
nvprof ./cling -xcuda to profile your interactive application.
This docker container can be used to
experiment with Cling’s CUDA mode.
Planned future developments for the CUDA mode include: Supporting of the complete current CUDA API; Redefining CUDA Kernels; Supporting other GPU SDK’s like HIP (AMD) and SYCL (Intel).
Conclusion
We see the use of Interactive C++ as an important tool to develop for researchers in the data science community. Cling has enabled ROOT to be the “go to” data analysis tool in the field of High Energy Physics for everything from efficient I/O to plotting and fitting. The interactive CUDA backend allows easy integration of research workflows and simpler communication between C++ and CUDA. As Jupyter Notebooks have become a standard way for data analysts to explore ideas, Xeus-cling ensures that great interactive C++ ingredients are available in every C++ notebook.
In the next blog post we will focus on Cling enabling features beyond interactive C++, and in particular language interoperability.
Acknowledgements
The author would like to thank Sylvain Corlay, Simeon Ehrig, David Lange, Chris Lattner, Javier Lopez Gomez, Wim Lavrijsen, Axel Naumann, Alexander Penev, Xavier Valls Pla, Richard Smith, Martin Vassilev, who contributed to this post.
You can find out more about our activities at https://root.cern/cling/ and https://compiler-research.org.